IT and Informatics Standards
Version 2.1—July 2023
Authors: Monica poelchau, Rob davey, Kim pruitt, nick salmon, Keith A. crandall, cibele sotero-caio, peter harrison, emillio righi, mark blaxter, ann Mccartney, tong wei, juan carlos castilla rubio, stephen richards, vincent (xiaofeng) wei
A.Overview
Coordinating the collection, integration, standardization, analysis, archiving and sharing of genetic data and related metadata is the core mission of Earth BioGenome Project (EBP). The EBP aims to undertake systematic sequencing and analysis of genomes from all known species, through collaboration with different institutes or groups around the world. Thus, EBP serves as a platform to connect biologists/genomicists within the EBP community and other stakeholders to foster collaboration and data sharing, in line with access and benefit sharing of the Convention of Biological Diversity Nagoya Protocol and related international agreements and regulations (see Access and Sharing Policies section). In this regard, the IT/informatics subcommittee needs to define minimum requirements, standard operating procedures (SOPs), recommended practices, and infrastructure requirements to support data and metadata handling, whether it is the collection of voucher specimen information, documenting the processes of data analysis, or providing access to data outputs. These informatics standards facilitate the EBP and its members to contextualize and reuse the genomic and associated meta-data collected as part of this international effort. By following standards, we expect to develop a model and build a platform which covers the infrastructures, production systems, archive repositories and analysis pipelines to manage the overall progress and data of the EBP more efficiently, to comply with the data sharing regulations and/or laws of different countries and political entities, and to promote sharing, mining and application of the data.
Figure 1. Establishing an IT/informatics standard framework with best practices and recommended resources to support the EBP/EBP Affiliated Project and other subcommittees.
A1. Access and Sharing Policies
The recommendation is to support the FAIR (Findability, Accessibility, Interoperability, and Reuse) principles) and place no restrictions on data access by submitting consensus reference genomes and corresponding sample metadata to INSDC (International Nucleotide Sequence Database Collaboration) managed data repositories. However, considering the Nagoya Protocol or related regulations in different countries and political entities, under certain circumstances the submitted genomics data may include a comment that links back to the submitters website for more information about restrictions/licenses but the comment may not include the license information itself. INSDC data repositories do not enforce licenses. In-country data generators are encouraged to work with an INSDC member to establish a workflow to broker submissions. The EBP recognizes that some researchers need to abide by nation-specific data policies and may need to establish local plans for how to meet the goals of data sharing.
A2. Data and Metadata Standards
In the archiving and sharing of omics data, most of the international public archiving systems and databases, such as those managed by the INSDC, already reflect community data format standards through the archiving process, data structure and data management practices. Genome sequence, annotation, and sample information standards should be compatible with those already established by the INSDC. Considering that the field of biodiversity research involves different species groups, these existing data schemas can be extended by working with INSDC members to supplement the existing standards if needed.
A3. Analysis and Pipelines
Considering different infrastructure capabilities and data analysis running environments across EBP partners, analysis pipelines will inevitably be varied, which may directly affect reproducibility. We should establish strategies for standardization and sharing of pipelines using descriptive and controlled workflow languages, versioning systems, and code repositories.
A4. Portal and User Services
EBP includes different research communities who may be using different standards or specifications or tools for their data analysis pipelines, data schemas, data archiving, and data sharing. A EBP central user services portal will provide useful services to these research communities in tracking and sharing this information, thereby making it easier for researchers to manage and use data files and pipelines.
B. BEST PRACTICES AND RECOMMENDED RESOURCES:
B1. Access and Sharing Policies
Compared to human genetic resources, there are fewer restrictions on data sharing of biodiversity genomic data, but some countries and regions have specific biodiversity protection or management laws and regulations. Therefore, under the premise of aligning to the Nagoya Protocol where appropriate, EBP can provide a more inclusive design to promote the data sharing of the entire project.
B1.1. Repositories
The EBP involves many types of metadata associated with the targeted genomic data, including information on the biological sample, sample vouchers, and even images. Although most of this can be archived by using existing international public biological repositories and the management systems of biobanks and museums, it is still necessary to clarify what types of data we need to store at every stage. Data (such as sequences and images) and metadata that are submitted to a repository should result in a persistent identifier for public citation (e.g., accession, digital object identifier (DOI)).
B1.2. Sharing policies and licenses
Figure 2. If in future EBP has its own data service portal, researchers can easily choose the appropriate license for the DMPs (Data Management Plan) based on best practices and recommendations made within the portal.
Unrestricted access to data and analysis tools and software is the main recommendation of the EBP. Much of the data and metadata generated by the EBP will be housed in INSDC repositories, where data sharing is subject to the INSDC data policy. In general, records in INSDC repositories are available for free and unrestricted re-use, while citing the original submission based on scientific best practices. Based on this policy, a specific separate license is not recommended for submissions to the INSDC. For submissions to other public repositories, we recommend the use of CC-0 or CC-BY. We acknowledge that this may not meet all of the situations and sometimes there are other factors that need to be considered. For example, the biodiversity law of Brazil allows conducting research with Digital Sequence Information (DSI) derived from genetic resources, but registration is required for publication or patent application for DSI users (*Silva MD, Oliveira DR. The new Brazilian legislation on access to the biodiversity (Law 13,123/15 and Decree 8772/16). Braz J Microbiol. 2018;49(1):1-4. doi:10.1016/j.bjm.2017.12.001). For these cases, other common licences are an option, such as the Creative Commons suite of licences (see Appendix 1). If in future EBP has its own data service portal, researchers can easily choose the appropriate license for their own project (Fig. 2) based on suggestions made within the portal.
B1.3. Survey - EBP Data-Sharing & Management (EBP ELSI Committee)
The Earth BioGenome Project (EBP) is an audacious endeavor to obtain whole genome sequences of representatives from all eukaryotic species on earth. In addition to the Project’s technical and organizational challenges, it also faces complicated ethical, legal, and social issues. The members of the EBP’s Ethical, Legal, and Social Issues (ELSI) Committee are building a robust, and inclusive data sharing and management ecosystem for the Earth BioGenome Project (Ethical, legal, and social issues in the Earth BioGenome Project. Proc Natl Acad Sci U S A. 2022 Jan 25;119(4):e2115859119. doi: 10.1073/pnas.2115859119).
Remark: The content below comes from Dr. Ann Mc Cartney's presentation at the IT and informatics workshop on March 31, 2022, and if you are interested in the detailed progress of EBP data sharing and management policies, you can keep up with the work of the EBP ELSI Committee(Fig. 3).
B1.3.1. Overview
Section 1 : Data-Sharing Policies/Ethos in place
Objective: To understand affiliated partners' data sharing policies in place, and to identify any data sharing regulations stipulated by their funding body/bodies.
Section 2 : Sample Collection
Section 3 : Indigenous Partnerships
Section 4 : Data Use
Section 5 : Data Release and Storage
Objective: To understand the if, when and where data generated by affiliated partners are being released, and where it is being stored.
Section 6 : Co-authorship Publication Policy
Section 7 : Policy Adoption and Future Development
B1.3.2. Respondents
30 responses, as of April 1, 2022, from :
European Reference Genome Atlas
Aquatic Symbiosis Genomics project
Darwin Tree of Life project
Soil Invertebrate Genome Initiative
Plant GARDEN
CanSeq150 and Canadian EBP
Fish 10,000 Genomes (Fish 10K)
California Conservation Project
Ag Pest Initiative
AfricaBP
i5K Initiative
Open Green Genomes
Genomics for Australian Plants
AusARG Reptile
Lilioid Monocots Core Group Genome Project
G10K
Global Genome Initiative
Euglena International Network
10KP Project
Dresden HQ Genomes Project
B10K
Endemixit
Squalomix
Threatened Species Initiative
Butterfly genomes project
Epizoic Diatom Genome Project
The Global Invertebrate Genomics Alliance (GIGA)
Diversity Initiative of the Southern California Ocean
The Catalan Initiative for the Earth BioGenome Project (CBP)
The Ungulate Genome Project
B1.3.3. Recommendations
Overall it may be difficult to enforce Affiliated Projects to sign the official EBP Governance Documentation with an enforced EBP Data Sharing Policy, due to institutional, federal and national regulations and legislations.
We can propose that all EBP Affiliated Partners are expected to read our EBP Data-Sharing & Management Best Practices including a section addressing Indigenous and Local Community Data-Sharing & Management Best Practices and apply them, to the best of their ability, when completing a EBP Data Management Plan. All projects could be expected to provide DMP.
These DMPs could be used 1) to assess our prospective Affiliated Projects as well as our current Affiliates and 2) to inform future EBP policy and standards development.
B1.3.4. Timeline
Figure 3. The timeline of the survey.
C. Data and Metadata Standards
C.1. Data and Metadata collection
The Earth BioGenome Project will generate massive amounts of data across the tree of life. The EBP’s overarching goals require that these data are accessible and re-usable by both humans and machines (The FAIR Guiding Principles for scientific data management and stewardship). With smaller datasets and analyses, researchers can pull the information that they need that is not described in the metadata record from additional text (usually associated publications, which are not always open-access). The large scale of the Earth BioGenome Project data will require public, preserved, structured, relevant, machine-readable metadata for effective re-use by the scientific community. The recommended data repositories in Table 1 provide mechanisms for metadata entry and storage. These general-use repositories necessarily require minimal metadata for the data types generated by a standard genome project; here, we encourage EBP members and affiliated project networks to provide a high standard of metadata beyond the minimum requirements.
We also recognize that collecting and reporting metadata can be onerous for the data submitter, in particular if it is dubious how and whether other scientists will (or even should) use it. How much metadata is enough? Will the required metadata sufficiently capture the complexity of the underlying methodology or experiment? In some projects, where the data are generated by only a few providers, rich structured metadata can be readily harvested; however, this becomes a challenge in a distributed project such as the EBP. Recognizing these challenges, we provide concrete and practical recommendations that we hope will allow a consistent and reasonable approach to metadata across large swaths of the project. Each affiliated project network should strive to implement a consistent approach to metadata within their network. See NCBI Bioproject PRJEB42239 www.ncbi.nlm.nih.gov/bioproject/687470 for an example implementation of genome metadata.
While we anticipate many different use cases for the EBP data, we expect the primary data generators and users to be in the realms of bioinformatics and biodiversity informatics. This expectation influences our recommendations.
Recommendations:
Incorporate metadata collection and entry into your project planning.
Why? Experimental details are more likely to be remembered and recorded accurately when the experiment is performed, rather than after the fact.
How? Use a platform that brokers submissions to an INSDC repository. For example, the COPO project and CyVerse platform provide mechanisms for metadata entry. Sample- and project-level metadata can be collected (and published) early in the project.
Collect the metadata for your project with future scientists in mind.
Why? The data generated by the Earth BioGenome Project should be re-used by future scientists. If relevant experimental details are only reported in published articles behind paywalls, then the data can’t be effectively re-used by all.
How?
Sample metadata: We are in the process of generating a EBP checklist for sample metadata, based on The Darwin Tree of Life Project’s metadata entry forms. These are designed to capture the most relevant metadata for this large-scale project, and following their templates will help ensure that your metadata is reasonably well scoped. However, if your metadata needs for your project differ substantially from the EBP recommendations, other INSDC sample checklists (e.g www.ebi.ac.uk/ena/browser/checklists or www.ncbi.nlm.nih.gov/biosample/docs/packages/) are perfectly acceptable, as long as as many metadata fields are completed as is possible and reasonable.
Sequence: Submission of sequence data to any INSDC repository should provide sufficient access to the data. Use of data brokers for submission to INSDC repositories is also acceptable. The sequence submission should be linked to the Earth BioGenome umbrella BioProject number, PRJNA533106 and, as relevant, to an additional BioProject representing an affiliated network. Sequence, assembly, and sample data should be submitted to the same INSDC center; e.g., do not submit assembly and sample metadata to one INSDC center and the assembled genome to a different center..
Assembly: Submission of genome assembly data to any INSDC repository should provide sufficient access to the data. Some projects may choose to provide additional information, structured or unstructured, about the assembly analysis details in other repositories, such as Zenodo. When choosing a repository, consider the longevity of the repository, and whether the metadata can be bidirectionally linked to the assembled genome sequence in INSDC and accessed by humans and machines.
Variants: Genetic variant data can be submitted to a center that brokers submissions to INSDC or directly to EMBL-EBI or DDBJ as NCBI no longer accepts submissions of non-human variant data. The preferred format for variant data is VCF (variant call format).
Project: Submission of project or study data to any INSDC repository should provide sufficient access to the data. We recommend generating an umbrella project, under the EBP umbrella project,for each EBP affiliated network.
Voucher: We recommend that EBP projects submit voucher data and metadata to an appropriate national repository (e.g., GGBN member repositories, http://www.ggbn.org/ggbn_portal/members/table) and include voucher IDs when submitting sample metadata. We will supply more specific recommendations in phase II.
Image: We recommend the Audubon core standard for multimedia resources (https://www.tdwg.org/standards/ac/), and Zenodo for archiving the images and receiving DOIs.
Given the existing fields in the metadata entry form, be as complete as possible.
Why? Attaching rich, structured metadata to a dataset is one of the best ways to make your data re-usable by others.
How?
Decide on your metadata forms in advance, and collect the metadata as you retrieve it. If you are working with a commercial sequence provider, ask in advance whether they can provide you with the necessary standard of metadata.
Use ontology or controlled vocabulary terms for your entries to the extent possible. EBI’s ontology lookup service is a great resource for this. It is often helpful for EBP networks to discuss and recommend ontologies that can be used for a particular domain or taxonomic group, given the large number of options available.
If you cannot supply sufficient information in the designated metadata fields, use a description field. In some cases, the existing repositories do not allow for sufficient granularity of your metadata for others to re-use it, i.e. the “bare minimum”. In this case, repositories will often provide additional fields where free-text is allowed. Providing detailed information there will allow future scientists to better understand your data, without having to access a manuscript. Again, it is helpful to include ontology terms or descriptions in these fields, for example the DTOL metadata schema suggests the use of the Environment Ontology (ENVO) for describing the habitat of collected organisms.
C.2. Best Practices - Genome Notes (DToL)
C.2.1. Introduction
The call from the Earth BioGenome Project (EBP) to “sequence everything” is leading to a very rapid increase in the number of high quality genome sequences being generated for all branches of the tree of life. Genomic science and its applications thrive and build on open data. Currently the process of formal publication lags significantly behind the pace of generating the genomes and current open-access models cost thousands of dollars/pounds/Euro per paper. When compared to the cost of sequencing, publication fees put a brake on FAIR open science.
At the Sanger Institute, the DToL team is building and championing a new mode of publishing genomes (Launching the Tree of Life Gateway, Threlfall and Blaxter 2021; DOI: 10.12688/wellcomeopenres.16913.1). Its vision is to produce, for each genome sequenced, a short paper - a Genome Note - that credits the individuals involved, makes the underlying metadata evident, and provides a formal assessment of the assembly quality, and also serve as formal statements of the open availability and reusability of the sequence.
C.2.2. Writing a Genome Note
Genome Notes cover the essential methodologies and biological information of the sequences. So, the submitter must describe the origin of the specimen that was used for sequencing; detail the methods used to extract and sequence the genetic material; and relay the bioinformatic processes that were used to assemble and fine-tune the genome sequence.
Authors can also include descriptive statistics and figures, which helps demonstrate the quality and accuracy of the sequences. Where applicable, authors should cite and summarize any previous publications that use the sequences presented.
Submit genome data
Assembly
Raw reads
BlobToolKit processing
Metrics on assembly (N50, % in chromosomes, etc)
Map raw reads -> coverage assessment
BUSCO -> biological completeness
Export XML for manuscript
Technical tables and figures from BTK
Add introduction
Add specimen image
Add methods
Submit to Wellcome Open Research
This DToL Gateway publishes Genome Notes on Wellcome Open Research that describe the genome of each species sequenced by the Darwin Tree of Life Project.
Data Note: https://wellcomeopenresearch.org/gateways/treeoflife
C.2.3. Universal Genome Notes
For publicly available data
A cloud based engine that “Write” the core metrics part of the MS
Emits
XML (text, figures (with hyperlinks), tables)
A checksum that validate the XML
Allow authors to add introductions and methods
Facilitates direct submission to F1000 and other journals
All Genome Notes will be hosted in the new Genome Sequencing gateway on F1000Research.
C.3. Best Practices - Next-generation germplasm digitalization (BGI-Research & WUR-CGN)
C.3.1. Workflow of the project
Figure 4 : Drafted a Consortium Standard for Lettuce Digitalization
C.3.2. Lettuce DataBase
Germplasm module: link to ex situ seed bank; develop a core collection.
Data module: generate processed datasets via an automatic standard pipeline.
Application module: ‘gene interact’ includes cis-regulation, protein interaction, genetic interaction.
The first version of the Lettuce Database: db.cngb.org/lettuce
Figure 5 : The design of the Lettuce database
D. Analysis and Pipelines
D.1. IT infrastructure
The EBP infrastructure requirements are large, not only for the final genome assembly data that needs to be archived, but also the storage and computing resources needed for intermediate analyses. We recommend the development of a mechanism to share the existing infrastructure capabilities for each affiliated project (Fig. 6).
D.1.1. Local resources
When considering the sharing of IT hardware resources in the various EBP affiliated projects, we recommend aggregating links to the different infrastructure resources which can be shared into a “hardware resources list”, and keep it regularly updated. If any groups encounter the lack of hardware resources which cannot be carried out, then they can find the resources from the “list” and use the internet or expressway to transfer data to other centers for analysis.
Figure 6. If in future EBP has its own data service portal, researchers can easily apply for the hardware resources in a “List” which are shared by the EBP affiliated project networks .
D.1.2. Cloud resources
D.1.2.1. Commercial cloud
If local and shared computing resources cannot meet the demand of EBP computing needs, elastic commercial cloud resources can be a suitable choice to complement local computing resources and provide on-the-fly expanded resources for targeted use. Cloud resources can be purchased on demand and expanded elastically, though the overall cost can be higher than local compute and storage. There are low-cost services such as offline storage, e.g., Amazon Glacier, but transmission of data into and out of these services can incur a significant cost. If your requirement for resources is temporary and your analysis workflows are well understood, cloud services are suitable and much cheaper than building new hardware facilities due to their pay-for-what-you-use pricing structure.
With respect to security, cloud services, such as Amazon Web Services (AWS), Microsoft Azure, and Google Cloud Platform (GCP) are designed with high security in mind and are kept iteratively updated. However, users of cloud computing service provider’s should confirm that such use is in line with the laws and regulations of the countries or regions from which the data are generated. Similarly, security of cloud resources supplied by these vendors is often left to individual users, so care should be taken to ensure data integrity and system security.
D.1.2.2. Self-built cloud
For institutions or groups with an IT team and suitable underlying hardware in managed data centers, an open source cloud framework like Openstack can be used to build a cloud service that can then be supplied to fulfill the same requirements listed in 3.1.2a.
D.2. Security (Backup)
Large international omics data repositories, such as INSDC and CNGB, already have mechanisms for data backup and security. Unless there are extenuating factors, there will be no data loss. But many times, when data is produced, they may not be archived into the data submission repositories in a timely fashion, and often there are no data backups locally. Considering the huge cost of genome sequencing projects in the EBP, it is required to archive data as soon as possible (Fig. 7).
At present, if a scientific research project, e.g., ICGC, can be added to the scientific data lists of some cloud service providers such as the Open Data project of AWS, the space for storing data can be free of charge.
If not, this will need to be discussed within the EBP partner project meetings to set out processes for data security and backups.
Figure 7. The large international omics data repositories, like INSDC, are the first choices for data backup.
D.3. Code repository
Although open access data are the basis of ‘omics research, the repeatability of data analysis pipelines is also critical for the accuracy and reliability of final research conclusions. In addition to the differences in experimental methods and sequencing platforms, the differences between analysis pipelines, algorithms and parameters directly affect the accuracy and quality of the data, and downstream interpretation.
We recommend that each EBP project establish a publicly accessible code repository, which can be Zenodo, or Github/Gitlab, to version, archive and manage the code of the analysis pipelines (Fig. 8).
Figure 8. The management and sharing of the pipelines. **The standardized and dockerized methods by WDL and Docker are not mandatory, but recommended options.
D.4. Workflows
Data processing workflow languages, like WDL (Workflow Description Language), can be used to standardize pipelines. By storing this descriptive specification about the tools used to complete an analysis in a suitable repository, we can ensure that the pipelines are easier to be used and maintained. For example, protocols.io can house descriptions of laboratory or analytical processes to aid reproducibility. Actual workflow description files can be stored and versioned in code repositories such as Github or GitLab. Therefore, we recommend that assembly and variant calling pipeline tools, parameters, and any called scripts are standardized and shared accordingly.
D.5. Containerisation
To aid reproducibility and reuse of data, EBP may provide online analysis tools and infrastructure to ensure the consistency of redeployment of analysis pipelines. There are suitable container technologies available that we can use to provide consistent pipelines in the later phases. We recommend Docker or Singularity images to contain any scripts, software and workflows. Bear in mind that DockerHub has recently stated they will delete Docker images that have not been used or updated within a 6 month time frame, so other mechanisms of persisting and sharing containers may be required.
E. Portal and User Services
E.1 Design - the data services portal
There are many biodiversity networks or groups under the EBP. Each group has its own strategy, which involves the design of scientific research topics, application of funding, selection of samples, selection of sequencing platforms, data analysis and data sharing. If we want to follow the progress of the different affiliated projects, we can not get the details from different data repositories, coordinate different IT infrastructure resources, and common data/sample processing issues easily. So a unified portal is necessary, though it needs to be designed and developed. Below are some basic functions and ideas for such a portal (Fig. 9.):
Figure 9. The basic design of the EBP data portal (which is in need of development) and its basic services.
E.1.1. Progress
Registers can update the progress of their projects, and every register can check the progress of all projects, including active querying of particular species and target species overviews for the entire project.
E.1.2. Exploration
Since different groups in EBP deposit data into different repositories, like NCBI SRA, EBI ENA or local storage, an EBP portal should allow for linkages across storage locations for quick and unified access to genomic data across the EBP. Such access and linkage would require a minimal metadata set that can be established based on the pre-defined vocabularies of different data repositories and indexed by the query engine.
E.1.3. Tools
While it would be impractical to store tools for genomic research in the EBP portal, we can provide links to effective tools and registered pipelines for analysis consistency and repeatability. Users can directly click and jump to the tool home pages through the outlink in this portal.
E.1.4. Brokering
In addition to the basic functions mentioned above, if we can design a component based on an integrated and comprehensive EBP data standard, at the same time collect the required information of each process in the production system (sample management system or lab management system), and also can be an agent or gateway to broker the data to EBI or NCBI, which would be very useful for the EBP and other genome research projects. The DTOL project has started such an integrated systematic approach involving open source data and metadata management tools as well as internal LIMS integration, and this model could provide insights into future EBP partner projects and their implementation of these recommendations.
E.2 Best Practices - Sample Tracking Systems (DToL)
E.2.1. Introduction
EBP partners will need to consider how they track the progress of samples as they move through the genomics pipeline. This has two main purposes:
It allows the EBP partner to manage their own sample management, sequencing and assembly processes efficiently;
It generates standard metrics and progress updates that can be escalated to EBP and combined with similar data from other partners to provide an overall assessment of EBP progress.
E.2.2. EBP Progress Metrics
To comply with EBP standards, all partners should be capable of reporting monthly on core metrics for EBP genomics (Table 2).
The EBP portal should include a front-end Dashboard with live, updated summary statistics across the metrics summarized in Table 2. This allows for accurate updating across the project and helps avoid duplicative genome sequencing efforts. Duplicate genomes should be allowed where appropriate metadata warrant such efforts.
E.2.3. Case Study - Wellcome Sanger Institute’s Tree of Life Programme
The Wellcome Sanger Institute’s (Sanger) Tree of Life Programme (ToL) is leading two EBP-affiliated projects: 1) the Darwin Tree of Life (DToL) project which aims to sequencing all 60k identified eukaryotic species in Britain and Ireland; and, 2) the Aquatic Symbiosis Genomics (ASG) project, which aims to sequence 500 symbiotic systems (>1000 genomes) from across the world’s freshwater and marine ecosystems. The collection of samples is through biodiversity partners, and DNA extractions are carried out within ToL. ToL works with the Sanger Institute’s Scientific Operations core to make sequencing libraries and generate raw sequence data. These data are assessed, assembled and curated by ToL teams before assemblies are submitted to the European Nucleotide Archive (ENA) and the International Nucleotide Sequence Database Consortium (INSDC). To support the receipt, storage and management of the tens of thousands of samples for these projects ToL has launched an IT systems project called Sample Tracking Systemisation (STS). The goals of STS are:
Enable easy entry and storage of sample metadata against defined schema;
Support sequence data submission and metadata brokering with genome repositories;
Track the status of every ToL sample at any stage of the pipeline;
Manage multiple identifiers for each sample and track relationships between samples;
Link samples and metadata to quality control information (e.g., DNA extraction yield);
Automate decision making and processing based on metadata wherever possible;
Enable clear reporting of pipeline and project metrics;
Provide easy access to ToL data and pipeline metrics for partners.
A discovery phase for this project was run between May-July 2020 and the project is now in build phase with an estimated initial go-live of the end of October 2020.
STS is being built using common open source tools including:
Web applications using the React Javascript framework.
Python/Django for server-side code.
RESTful APIs.
Databases using PostgreSQL.
Message/event queues using RabbitMQ/Kafka.
Openstack.
STS also uses COPO (originally Collaborative Open Plant Omics portal, built by Rob Davey’s group at the Earlham Institute) to provide sample metadata collation, validation and data brokering support.
High level roadmap for STS:
Phase 1 (end Oct 2020)
Go live with support for sample metadata ingestion via COPO and storage in the STS database, plus early sample management, compliance management and work request generation. Go live of DToL public portal including project metrics and sample look-up.
Phase 2 (end Jan 2021)
Go live with support for production and R&D including laboratory information management systems (LIMS), data warehouse and electronic lab notebook integration. Will deliver sample tracking capability throughout sample management and production. Phase 2 will also deliver continuous improvements to the sample management capability provided in Phase 1.
Phase 3 (end Apr 2021)
Go live with support for assembly and curation including integration with informatics management tools and databases. Will deliver sample tracking capability throughout the entire Sanger Institute pipeline. Phase 3 will also deliver continuous improvements to the capabilities provided in Phase 1 and 2.
E.3. Best Practices - Genomes on a Tree (GoaT) (Tree of Life)
E.3.1. Introduction
Under the umbrella of the EBP, many initiatives will generate large numbers of reference genomes. The distributed nature of this work makes coordination essential to prevent duplication of effort and facilitate collaboration between projects. While public sequence databases hold data for completed projects, the EBP community would benefit from a centralized source of information about intent and progress of sequencing projects. In addition, the scoping and delivery of assemblies benefits from prior estimates of genome size and karyotype, but existing data are scattered in the literature.
To address these issues, the Blaxter Lab in capacity of the Tree of Life Programme has developed Genomes on a Tree (GoaT), an ElasticSearch-powered, taxon-centered database that collates observed and estimated genome-relevant metadata, such as genome size, karyotype and assembly metrics, for eukaryotic species. Missing attribute values for taxa are estimated from direct values using phylogenetic imformation. GoaT also holds declarations of actual and planned activity across genome sequencing consortia. Data such as priority lists and in-progress status, as well as availability of assemblies on public sequence archives belonging to the International Nucleotide Sequence Database Collaboration (INSDC www.insdc.org) can be queried and retrieved along other relevant metadata at taxon or assembly indexes.
GoaT has been adopted as a centralized search and metadata aggregator for the Earth BioGenome Project and currently reports priority lists from various projects in the EBP Network, including the Darwin Tree of Life project (focussed on the biodiversity of Britain and Ireland). The GoaT team is actively soliciting additional projects to share their progress and intent lists so that GoaT displays a real-time summary of the state of play in reference genome sequencing. Standard guidelines and ontology for lists submissions are under refinement to allow any project to make explicit declarations of intent and progress. Cross referencing to other data systems such as the INSDC sequence databases, the BOLD DNA barcodes resource and Global Biodiversity Information Facility- and Open Tree of Life-related taxonomic and distribution databases will further enhance the system’s utility. Further metadata, such as ploidy and sex chromosome systems are currently under curation, to augment the utility of GoaT in supporting the global genome sequencing effort.
Goals
Centralized source of genome-relevant metadata for the global community
Store and provide public access to plans and progress in genome sequencing at the Tree of Life programme at the Wellcome Sanger Institute, and to offer coordination across biodiversity genomics projects in the wider EBP network.
Prevent duplication of efforts
How is GoaT useful to EBP affiliates
GoaT can be used throughout all steps of a sequencing project: planning, execution, reporting, or even for data exploration. The versatility of GoaT queries allows EBP affiliates to retrieve lists of species and generate reports, so that resources are used efficiently. The GoaT announcement publication includes detailed use case examples of how GoaT can be used in each of these steps, with links to each query results. This information and other tutorials can also be found on the GoaT help page. Questions in each of these steps include:
Planning: Target Lists : What should we sequence first? What has been already sequenced? Are there taxonomic gaps that should be further explored for sequencing?
Progress: Genome size estimates : How much sequence should we generate?
Progress: Karyotype information : What will a complete genome assembly look like?
Reporting: Exploring EBP-affiliated project pages: What is the current state of sequencing for EBP and each of its affiliates?
E.3.2. Projects
Earth BioGenome Project
EBP Affiliated Project Networks
AFRICABP - African BioGenome Project (AfricaBP, PRJNA811786)
ARG - Australian Amphibian and Reptile Genomics Initiative Collaboration (AusARG)
CCGP - California Conservation Genomics Project (CCGP - PRJNA720569)
ERGA - European Reference Genome Atlas (Pilot project - PRJEB47820)
EUROFISH - The Euro-Fish Project at the MPI CBG (PRJNA393850)
LOEWE-TBG - LOEWE Centre for Translational Biodiversity Genomics (PRJNA706923)
METAINVERT - Soil Invertebrate Genome Initiative (PRJNA758215)
SQUALOMIX - Genome Sequencing and Assembly of Chondrichthyans (PRJNA707598)
E.3.2. How to retrieve information from GoaT
GoaT can be queried through a mature API (application programming interface), and It has a web front-end that includes data summary visualizations.
User Interface (Web) goat.genomehubs.org , screenshots below:
API goat.genomehubs.org/api-docs
Command Line Interface github.com/genomehubs/goat-cli
E.4. Best Practices - Data portals for Darwin Tree of Life (DToL) and European Reference Genome Atlas (ERGA)
The DToL partners have developed a cohesive end-to-end process to take organisms from the field through to publication of high-quality assembled and annotated genomes in the public domain. By collating specimen metadata and tracking information through identification, DNA barcoding, extraction, sequencing, assembly, curation, annotation, and submission, the process assures that genomes are published with rich, informative, and accurate descriptors.(From Sequence locally, think globally: The Darwin Tree of Life Project, www.pnas.org/doi/10.1073/pnas.2115642118)
Figure 10. An end-to-end process that assures data integrity, accuracy, and quality.
E.4.1. DToL data portal
Figure 10. Home page of the DToL data portal, URL : portal.darwintreeoflife.org
E.4.2. Framework of the portal
A single access point to the wealth of DToL metadata, raw data, assemblies and annotations.
Filter by data status and clade and search across all metadata fields. Static URLs for sharing.
Cross reference to additional sources of information, add common names, download metadata.
Main functions
Interactive Phylogenetic search.
Status tracking.
Public (often higher) level view of sequencing and annotation status of any species
Status triggers based on deposition of data into public archives (all records are linked) and via controlled API push updates to the portal.
Programmatic access API
Bulk download
Multiple annotation sources
For DToL Ensemble is providing rapid annotation of sequenced species, and will also conduct annotations for ERGA.
Portals also need to track and present community annotations.
Standardized identifiers and file format developments play a key role in this with DToL, INSDC and many others working on standardizing GFF3 annotation.
Multiple levels of portals
Species assemblies and annotations can and will exist in multiple portals.
Important to promote consistency in
Metadata data standards and data richness.
Metrics for data analysis
Provenance and licensing (data openness)
Interoperability of portals and databases
Data Provenance
Increases in importances as the scale of regions and component projects grows.
Need to clearly know the component/secondary project generated the data, under what standards, methods and metrics.
Require open data or clear recording of third party access restrictions and are propagated from source and machine readable.
Clear Nagoya protocols and access and benefit sharing.
Portals should provide filtering on all of these criteria. For example, displaying only open source data.
Need for data wrangling or a common data layer
At the scale of EBP (and ERGA) there is a need to handle multiple metadata standards, protocols and data formats..
Either:
Need agreement on a common data layer (API standards and/or data format) so that regardless of source the same data can be accessed and harmonized.
Effort in data wrangling to harmonize (preferably largely automated).
Issue of how to effectively present this data:
Merging similar data types, dealing with common names.
Smoothing descriptions for more cohesive experience
Dealing with gaps in data
Also will be a variety of quality, methodologies and QC to explain.
In general follow a philosophy of providing the metrics and letting a user decide which reference to use.
Technical scale
ASG, DToL and ERGA data portals are multi cloud deployments.
Key technologies are:
Database layer: Elasticsearch and Neo4j
Frontend Layer: Angular
Production deployment: Self healing & k8s server in Embassy Cloud and Google Cloud Run as fallback service
Graph based phylogeny utilizing the neo4j graph database (complex parent-child relationships ) is essential for rapid traversal of available taxa.
Can deal with simultaneous scale of users.
Can deal with scale of data assemblies and annotations of species.
E.5. Best Practices - Data Portal of Catalan Initiative for the Earth BioGenome Project (CBP)
E.5.1. Introduction
CBP aims to generate a detailed catalogue of the genome of the eukaryotic species of the Catalan Countries. The project is initially raised by the Societat Catalana de Biologia (SCB) and is developed jointly with the Institució Catalana d’Història Natural (ICHN), both subsidiaries of the Institut d’Estudis Catalans (IEC).
It currently serves as the umbrella for a wide network of expertise that extends throughout the territory and involves the collaboration of institutions, institutes and research centers, museums, zoos, botanical gardens, societies and expert entities in the knowledge and study of species, as well as powerful technological infrastructures for genomic sequencing and computing.
Therefore, the CBP is a collaborative project that has the support and direct participation of more than 20 centers throughout the Catalan-speaking territory, including Catalonia, the Valencian Country, the Balearic Islands, Andorra and Banyuls-sur-Mer (France).
E.5.2. (meta)data of CBP
Figure 10. CBP (meta)data flow
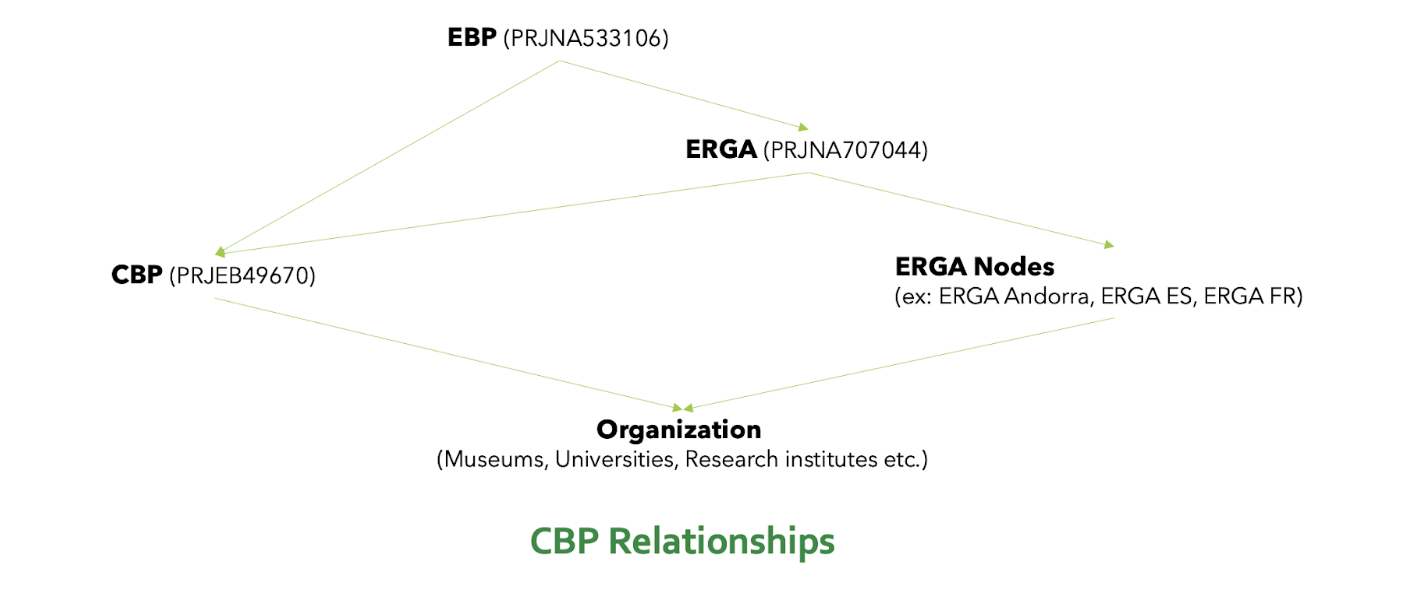
Figure 11. CBP relationships
E.5.3. Data(Biogenome) Portal
Integrare date from different sources
Submission status tracking
Local data submission
Data visualization
Phylogenetic tree
Geographical location
Project visibility
E.5.3.1. Tech stack
Multi-container web application:
Flask (python) REST API → Server
VueJs/Nginx (Javascript) UI → client
MongoDB → database
E.5.3.2. Main feature
The BioGenome Portal can be used to display data at any level of the EBP (and beyond).
Configuration
Portable
Flexible data integration system
Cronjobs
Admin UI
E.5.3.3. cronjobs
Scheduled background processes to retrieve data from:
NCBI (retrieve assemblies and sample accession) give a Bioproject ID
EBI/Biosample* given a list of project names
*from DToL data portal cronjobs
E.5.3.3. Admin UI
ENA reads/assemblies by sample accession
User input from admin area
Local Sample Form
form to submit sample metadata locally
Organism Image/Image URL
Organism local name/s
* The UI consumes the API
Appendix 1.
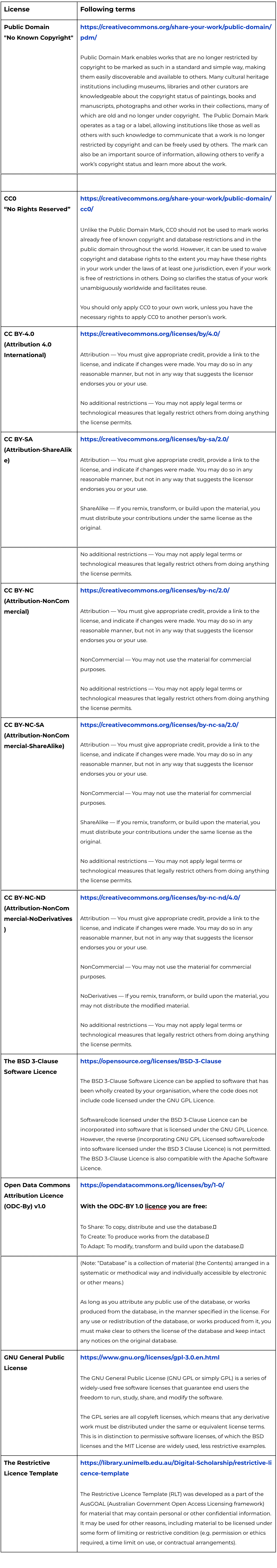
Reference: licenses
In EBP, we choose INSDC as the default repositories for researchers to share their datasets. But in order to ensure that the researchers get credit for the data or softwares, and follow Nagoya Protocol or related regulations in different countries and political entities, publishing without restriction is not always ideal. We have also enabled the option to publish under other licenses.
The following table can bring you some suggestions.